Vadym Ivanchuk
I'm a Biomedical Engineer committed to advancing precision medicine through a blend of computational biology, artificial intelligence, and medical engineering.
My main focus is on bridging the gap between research and clinical practice, translating scientific advancements into practical solutions that directly benefit patients. With expertise in bioinformatics and machine learning, I've spearheaded projects ranging from developing cancer bioinformatics pipelines to designing AI-based devices for multifaceted rehabilitation.
Feel free to explore my webpage to learn more about my past projects and contributions to the field.
Karolinska Institutet
 As a bioinformatician at the
Clinical Genomics Core Facility, I contributed
to the development and integration of genomic and transcriptomic computational tools into
clinical settings. My work focused on two key areas: designing bioinformatics solutions to
identify and interpret somatic mutations in cancer and automating bioinformatics workflows
to improve data management and delivery.
As a bioinformatician at the
Clinical Genomics Core Facility, I contributed
to the development and integration of genomic and transcriptomic computational tools into
clinical settings. My work focused on two key areas: designing bioinformatics solutions to
identify and interpret somatic mutations in cancer and automating bioinformatics workflows
to improve data management and delivery.
Spanish National Cancer Research Center
 I was a research assistant at the
Bioinformatics Unit. Advised by
Dr. Fátima Al-Shahrour, I contributed
to the optimization of a single-cell RNA sequencing pipeline and the validation of a
bioinformatics method aimed at linking subpopulations of tumor cells to cancer-specific
treatments.
I was a research assistant at the
Bioinformatics Unit. Advised by
Dr. Fátima Al-Shahrour, I contributed
to the optimization of a single-cell RNA sequencing pipeline and the validation of a
bioinformatics method aimed at linking subpopulations of tumor cells to cancer-specific
treatments.
Puerta de Hierro Majadahonda University Hospital
 During my internship at the
Liquid Biopsy Laboratory
of the Oncology Department, I worked with
Dr. Atocha Romero
on the bioinformatics analysis and molecular profiling of liquid biopsy samples from lung
cancer patients. This experience was crucial in shaping my future research path in cancer
genomics.
During my internship at the
Liquid Biopsy Laboratory
of the Oncology Department, I worked with
Dr. Atocha Romero
on the bioinformatics analysis and molecular profiling of liquid biopsy samples from lung
cancer patients. This experience was crucial in shaping my future research path in cancer
genomics.

Technical University of Madrid
 Master of Science in Biomedical Engineering.
It was during this program that I first got into bioinformatics. Mentored by
Prof. Dr. Enrique J. Gómez Aguilera,
I also coordinated a semester-long project aimed at digitalizing the monitoring of children
with cystic fibrosis at the
Ramón y Cajal University Hospital.
Master of Science in Biomedical Engineering.
It was during this program that I first got into bioinformatics. Mentored by
Prof. Dr. Enrique J. Gómez Aguilera,
I also coordinated a semester-long project aimed at digitalizing the monitoring of children
with cystic fibrosis at the
Ramón y Cajal University Hospital.
Biogipuzkoa Health Research Institute
 I spent a year as a research assistant in
Prof. Dr. Marcos J. Araúzo-Bravo's group
at the
San Sebastián Oncology Hospital,
leading the software development of a platform for collecting, analysing, and interpreting
cancer patient-reported outcomes (PROs), and closely collaborating with the clinicians of the
Medical Oncology Department.
I spent a year as a research assistant in
Prof. Dr. Marcos J. Araúzo-Bravo's group
at the
San Sebastián Oncology Hospital,
leading the software development of a platform for collecting, analysing, and interpreting
cancer patient-reported outcomes (PROs), and closely collaborating with the clinicians of the
Medical Oncology Department.
Technical University of Madrid
 I obtained an internship to work with the
Life Supporting Technologies
research group while completing my studies. During this time, I contributed to projects within
the university's
Smart House Living Lab,
focusing on domotics, telemedicine, and artificial intelligence for active and assisted
living.
I obtained an internship to work with the
Life Supporting Technologies
research group while completing my studies. During this time, I contributed to projects within
the university's
Smart House Living Lab,
focusing on domotics, telemedicine, and artificial intelligence for active and assisted
living.

Technical University of Madrid
 Bachelor of Engineering in Telecommunication Technologies and Services, with a minor in Electronic Systems. I focused my studies on embedded systems and machine
learning, exploring their applications in healthcare under the guidance of
Prof. Dr. María Teresa Arredondo.
Bachelor of Engineering in Telecommunication Technologies and Services, with a minor in Electronic Systems. I focused my studies on embedded systems and machine
learning, exploring their applications in healthcare under the guidance of
Prof. Dr. María Teresa Arredondo.
Projects
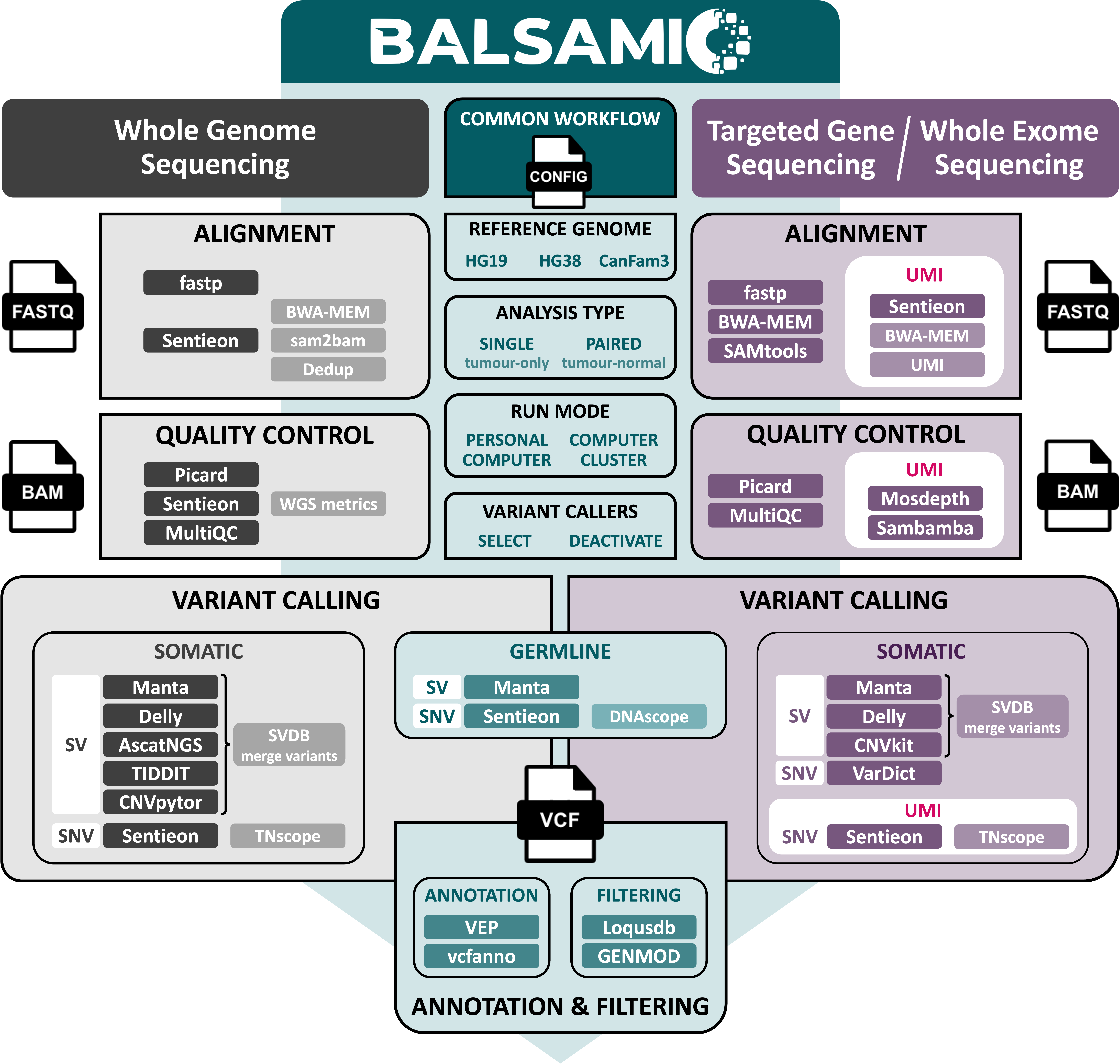
BALSAMIC
Bioinformatic Analysis Pipeline for Somatic Mutations in Cancer
BALSAMIC is a Clinical Genomics Core Facility effort aimed at facilitating the identification of somatic mutations and the interpretation of large-scale DNA sequencing data in cancer patients, paving the way for improved diagnostics and treatment.
It is a Snakemake-based configurable bioinformatics pipeline that integrates multiple somatic variant-calling algorithms to detect SNVs, InDels, CNVs, and SVs. BALSAMIC supports whole-genome, whole-exome, and targeted gene sequencing, enabling the processing of tumor-only and tumor-normal sample pairs, as well as assays with unique molecular identifiers.
I contributed to its development, maintenance, automation, and integration within the group's infrastructure, as well as addressed clinical visualization and interpretation needs.
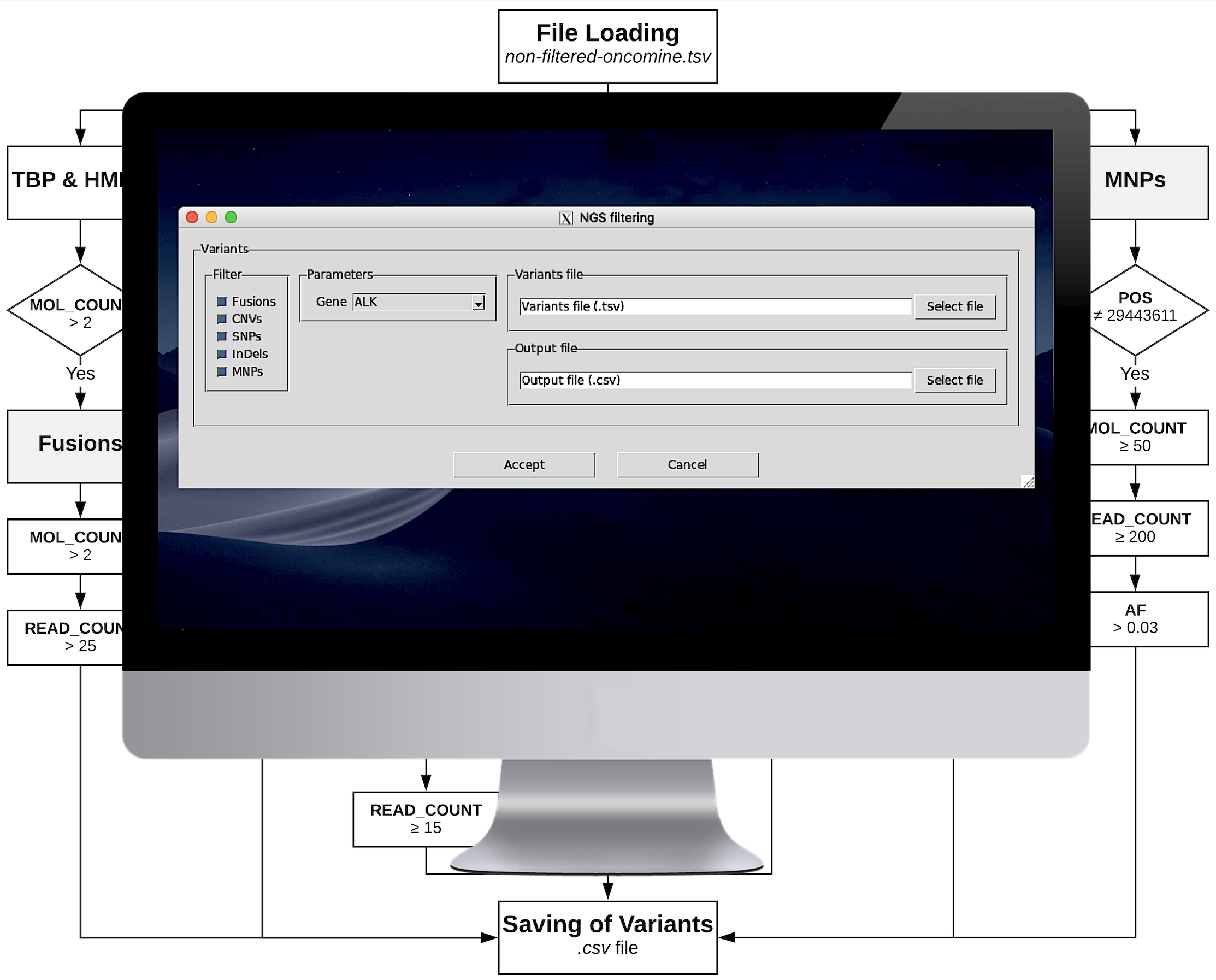
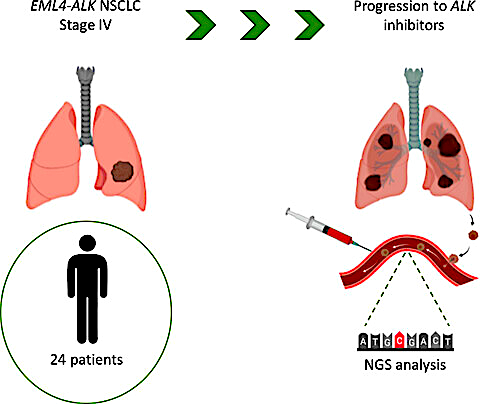
vALK
Bioinformatics Pipeline for the Detection of Somatic Mutations in Liquid Biopsy Samples from Non-Small Cell Lung Cancer (NSCLC) Patients
As part of my master's thesis project, I developed a somatic variant filtering pipeline and a user-friendly graphical interface with the goal of automating the manual curation of next-generation sequencing data conducted by researchers and clinicians at the Puerta de Hierro University Hospital.
This project involved integrating experimental and computational methodologies to identify actionable mutations in NSCLC patients, provide guidance for clinical treatment decisions, and improve understanding of the screening and diagnostic capabilities of liquid biopsies.
Some of the findings and methodologies resulted in a scientific publication, and currently, the developed algorithm is extensively used in clinical practice.
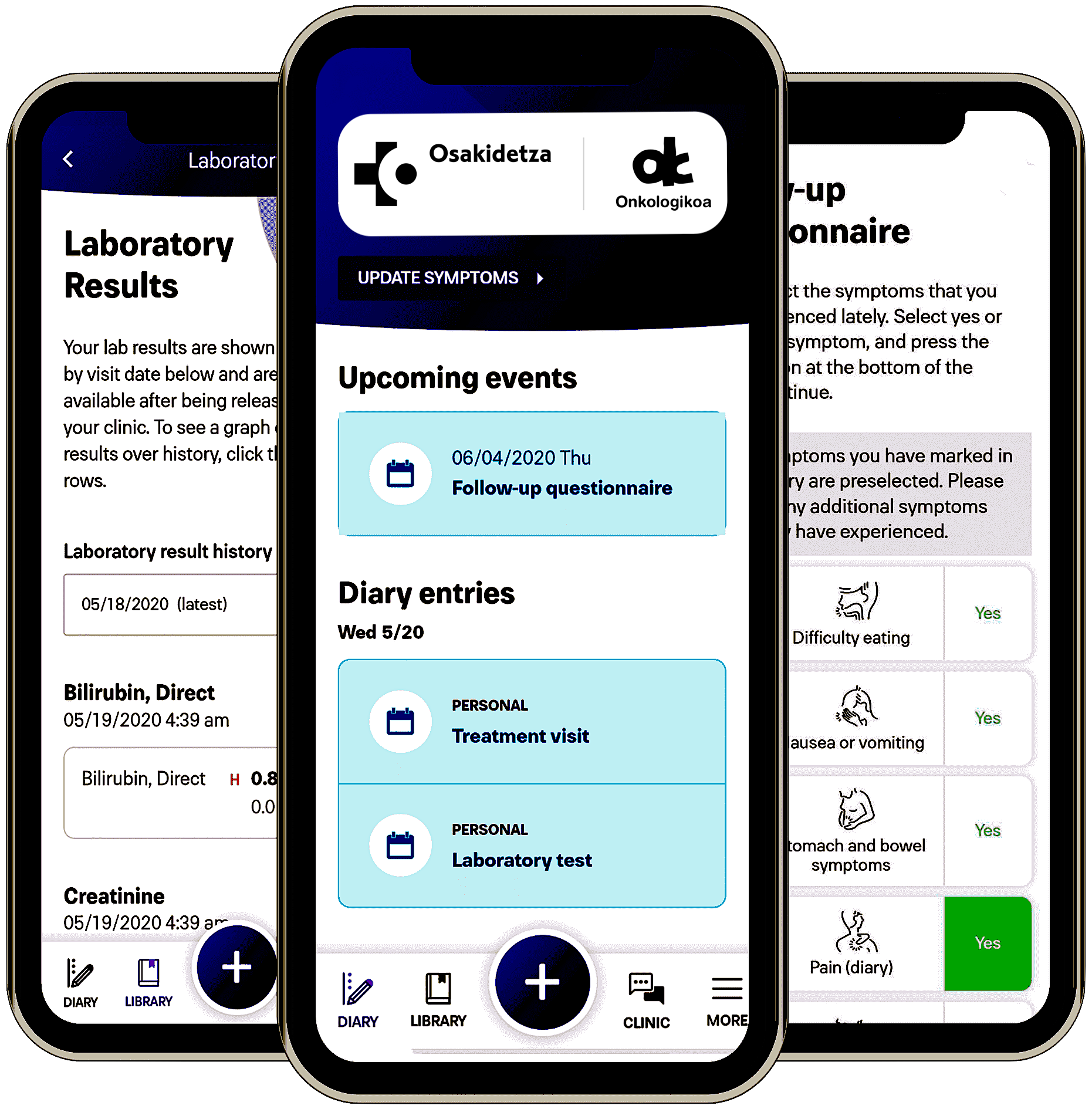
OnkoPROs
Platform for the Management and Follow-Up of Oncological Patients
OnkoPROs is an interoperable web, mobile, and server platform created to assist health professionals at the San Sebastián Oncology Hospital in tailoring treatment plans and remotely monitoring cancer patients undergoing treatment and rehabilitation, thus minimizing their frequent visits to the hospital for routine check-ups.
The platform includes a web application for clinicians to design and gather patient-reported outcomes (PROs), as well as a mobile application for patients. Through the mobile app, patients can complete questionnaires, report their health status, chat with their doctors, and receive the results of their clinical evaluations.
OnkoPROs was developed using Angular and NativeScript for the frontend, and Nginx and JavaScript for the backend, connecting to the medical record database of the hospital.

FERehab
Facial Expression Recognition Machine Learning Tool for Multifaceted Rehabilitation
FERehab originated from my bachelor's final project with the aim of enhancing the functional, emotional, cognitive, and social abilities of elderly individuals with Parkinson's Disease (PD).
It features an AI and computer vision-based real-time facial expression recognition model, along with an interactive game for patients, all integrated into a Raspberry Pi smart mirror. Currently, it's in the proof-of-concept phase at the Smart House Living Lab of the Technical University of Madrid.
On the technical side, the machine learning model has been designed using a Convolutional Neural Network (CNN) architecture, a Generative Adversarial Network (GAN) to expand the training dataset, and transfer learning methods.
Publications